Some visually appealing images acquired during our research
See also our 3D movies:
|
Cover art and graphical abstract from our review in Analytical Chemistry on Secondary Ion Mass Spectrometry: Characterizing Complex Samples in Two and Three Dimensions
|
|
Topography corrected, 3D visualisation of the membrane (green, m/z 184.1, phosphocholine) and nucleus (red, m/z 136.1, adenine) chemistry in the frozen‐hydrated HeLa‐M cells following the use of PCA to identify the interface between the cells and the substrate to allow more representative data reconstruction. The visualised area is the same as the analysis area in the SIMS experiment, 250 × 250 μm2, but inclined in order to aid 3D visualisation. |
|
Topography corrected, 3D visualisation of the membrane (green, m/z 184.1, phosphocholine) and nucleus (red, negative scores on principal component 5) chemistry in the fixed and freeze‐dried HeLa‐M cells following the use of PCA to identify the interface between the cells and the substrate to allow more representative data reconstruction. The visualised area is the same as the analysis area in the SIMS experiment, 250 × 250 μm2, but inclined in order to aid 3D visualisation. |
|
Distribution of lipid (DPPC; red), diacyl glycerides (DAG; green) and salts (blue) in a cross-section of rat kidney. 320 x 320 pixels = 9 x 9 mm |
|
Distribution of cholesterol and lipid (DPPC) in the white/grey matter boundary region of brain tissue by ToFSIMS.
|
|
By former student and PDRA Matt Baker for use by Analyst [journal article] |
|
Pseudo colour PCA images derived from TOF-SIMS of Streptomyces coelicolor on silicon (using 40 keV C60) |
|
ToF-SIMS 3D image of Xenopus laevis Oocyte, principal component 1 |
|
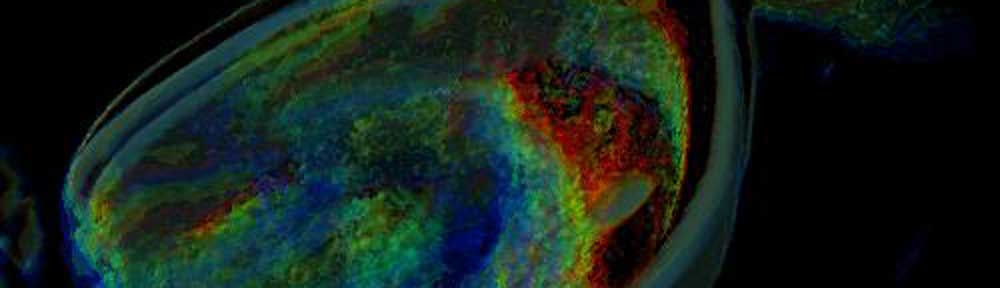
Distribution of Cholesterol in Xenopus Laevis Oocyte membrane. |
|
The original Ionoptika J105 3D Chemical Imager (click to enlarge) |
Images of specific ions present in HeLa (cervical cancer) cells (top) |
|
HeLa (cervical cancer) cells following maximum autocorrelation factor (MAF) analysis |
|
Human cheek cells
|